
Abstract
Results are presented of a genetic algorithm (GA) solving the Travelling Salesman Problem (TSP) with 20,000 city locations. Additionally, the performance of PMX vs OX1 vs “no crossover” schemes are compared, along with some thoughts for improving performance.
Hardware is that of an Intel i6700. Implementation is C# (.NET v5.0.1).
Introduction
A little while ago, I presented some initial results with a genetic algorithm solving the Travelling Salesman Problem (TSP) with 10,000 cities. That is a large number of cities.
I’ve done a little work on it since then and, here, I describe results with 20,000 cities along with some interesting observations regarding crossover.
The genetic algorithm is of my own devising and is implemented in C#. I am calling it EVOLGEN. It was developed for use in trading algorithms. The fact that it can also be employed to solve TSP merely allows for testing and comparison with other algorithms and approaches. I also found the subject of interest, and so persued it a little more than necessary.
What is a Genetic Algorithm?
A genetic algorithm is a search optimization technique which derives inspiration from biological notions concerning genetics and evolution. In short, we “breed” candidate solutions to the problem we want to solve and let them evolve. This is an approach used to train neural networks, but we can also apply it to the TSP.
Travelling Salesman Problem
The TSP, or “Travelling Salesman Problem”, is an NP-hard computational problem. In short, a “salesman” needs to visit a number of cities and obviously wants to find the shortest route between them. If the number of cities is small, say 10 or so, we can just look at them a draw a good route ourselves. However, as the number of cities becomes larger, the number of possible routes explodes according to a polynomial relationship.
20K Cities in Context
We can calculate the number of possible tours for n locations, where the tour is a closed path, as follows:
Rn = (n - 1)! / 2
For 20,000 locations, Rn is a staggering: 4.548 × 1077332
For comparison, the number of atoms in the Universe is estimated at a mere 1086 [1].
Now, searching the Internet, I can find references to other TSP projects dealing with up to 1,000 cities. I have found little reference to projects attempting 10K – 20K city points. Below are couple examples of what I did find. (Drop a comment, below, if you can provide any information regarding TSP with comparable city numbers.)
For example, there is a popular GA library for C# called GeneticSharp [2] for which animation is presented of it solving for 50 cities in around 10 – 14 seconds.
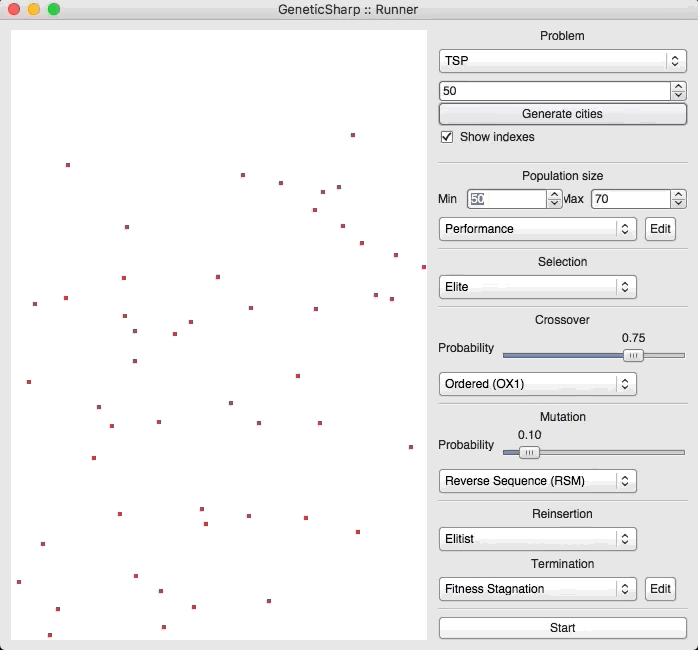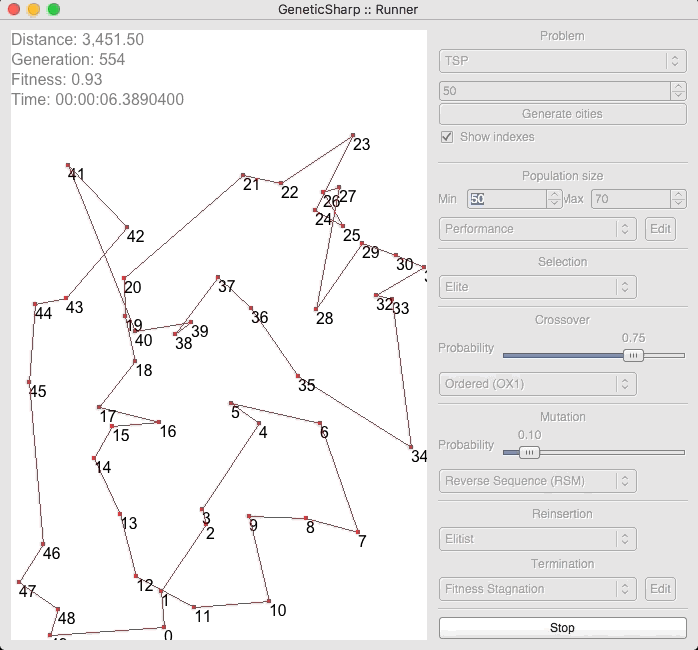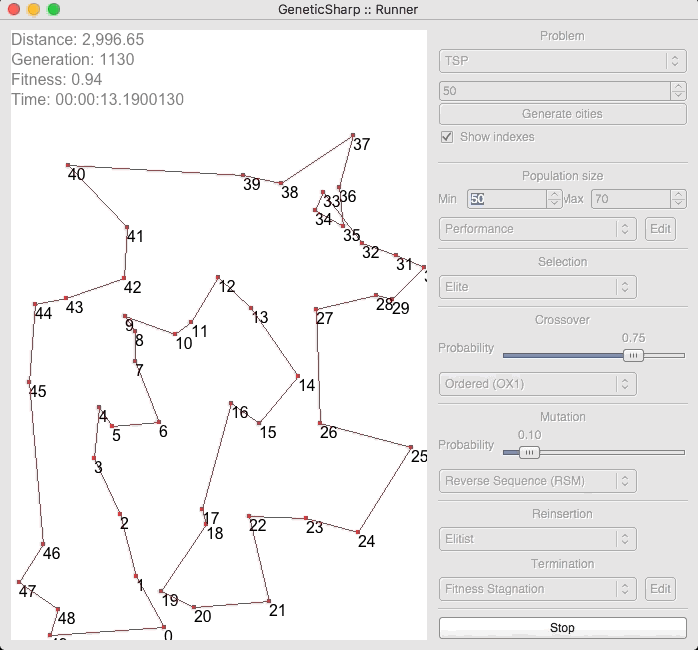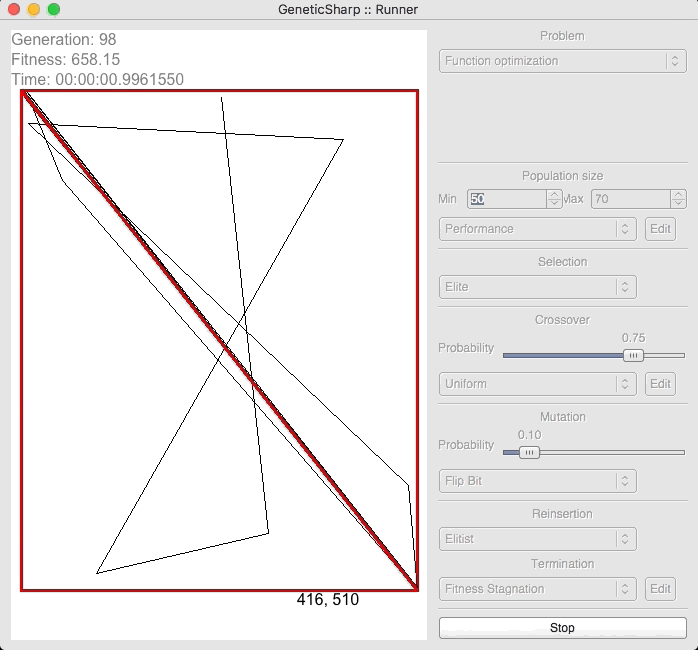
I’m presenting this because, when I initially ran EVOLGEN against 50 cities, it solved it nearly instantly (in under a second). This is what prompted me to test my own GA with a much larger numbers of locations.
I also found a video of a simulated annealing algorithm [3] solving for 1,000 cities. Simulated annealing is a different technique from that of a GA. The implementation referenced does a fast job — impressive in fact — with the video indicating that took around 1 minute 40 seconds to arrive at what it considers the solution.
The source code for this is on GitHub, although it links to a pre-built library. It is written in C++.
A Rather Surprising Result
Below, is a screenshot of the EVOLGEN “Test Rig” showing a 1,000 city result after 20 seconds. Here, it has essentially arrived a “satisfactory solution” but continued “fining tuning” the result after this. In this test, it stagnated after around 53 seconds. Performance is roughly comparable to the simulated annealing above (although, again, these are different approaches written in different languages).
In these screenshots, the route is shown on a grid with both X and Y in the range [-1.0, +1.0].
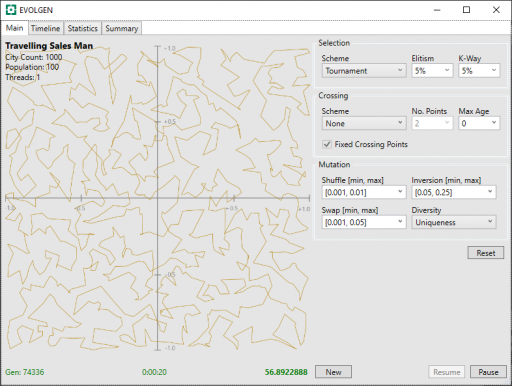
Now, if you take a close look at the screenshot above (click to enlarge), you will see the “Crossing Scheme” is set to “None“. What does this mean?
EVOLGEN supports the following “exclusivity” preserving crossover schemes:
- None
- PMX
- OX1
Both PMX [4] and OX1 [5] are commonly used schemes with TSP and preserve value exclusivity (it is necessary to ensure that each location remains visited only once and none are omitted). Notice that the GeneticSharp example animation, above, indicates that OX1 is used. In my initial experimentation, I was using PMX which tends to preserve more information than OX1, however, it is computationally more expensive.
In the “None” scheme, which was added because I thought it would useful only for test comparison purposes, no crossover actually occurs. Instead, we have a directed search, but one driven by random mutation only. The surprising result, which I found by accident, is that no crossover out-performs both PMX and OX1.
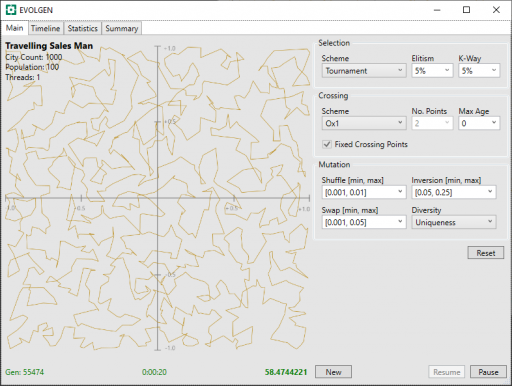
Below, are presented 2 screenshots, each taken after 20 seconds of TSP, with the PMX and OX1 schemes. Mutation parameters have been adjusted, by trial and error, for optimal results and a fixed seed has been used in the random generation of route locations (location are the same for all tests).
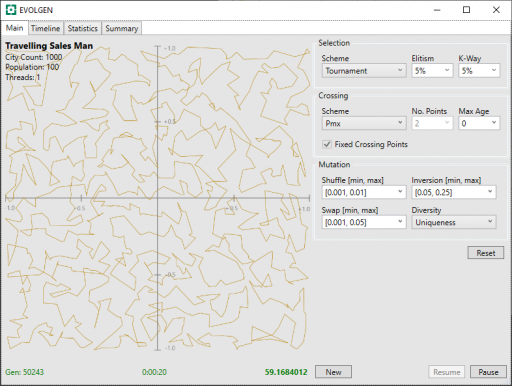
In these, above and below, the generation number is shown in the screenshot bottom-left, the time center, and the route distance bottom-right. Note that shorter route values are better.
We include the result from the “None” scheme, previous, and present these results are follows:
| Scheme | Generation (after 20 sec) | Distance (after 20 sec) |
|---|---|---|
| None | 74336 | 56.892 |
| PMX | 50243 | 59.168 |
| OX1 | 55474 | 58.474 |
In this limited test, both PMX and OX1 give similar solution distances after the allotted time, however OX1 processed more generations in that time. Because “None” was not encumbered with having to perform crossover at all, it was able to munch through more generations than the other two.
In short, per generation, PMX is the best out of these, but takes longer per generation. Ultimately, it is the time taken that we care about and, by this measure, it would seem that not performing crossover at all seems a good option.
Speculative Thoughts on Crossover
Performing no crossover appears to be a successful approach. I would surmise that, in preserving value exclusivity, both PMX and OX1 destroy much of the order information contained in the parent genomes when performing crossover, thus offsetting much of the advantage of their use.
There are other more complex crossover schemes, such as Edge Recombination. I have not implemented or tested this.
Now for 20,000 Cities
Because of the permutation explosion of TSP, running against 20K cities presents a far more formidable challenge than a mere 1000 locations. In fact, I ran into problems with my Test Rig app hanging because of the time taken to render the results on screen (I solved this by generating the image in a background thread).
Here is the problem faced at the start of the run. We can see that an early random route yields a monstrous tangle of solid color. How on earth can anything solve this?
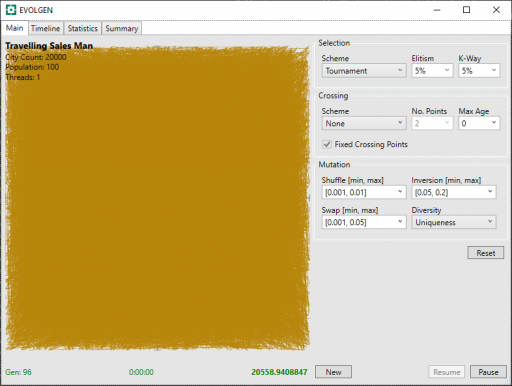
I also implemented a graphing component for use with this project and this shows us that progress is, indeed, being made.
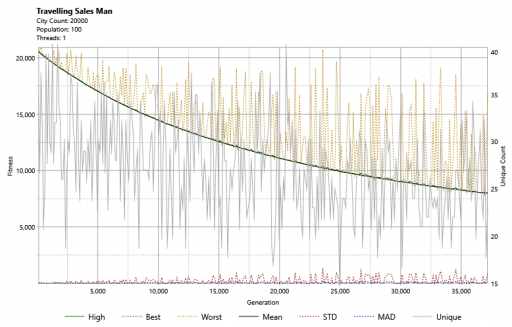
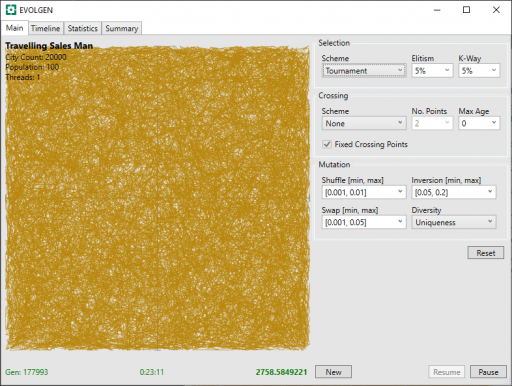
Clearly, this is going to take some time. After 23 minutes, below, we can see light appearing. We can also see that the distance (in green bottom-right) has decreased massively in this time: from 20558 to 2758.
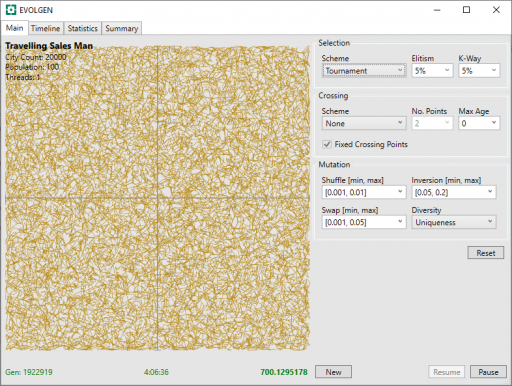
Additional progress now gets harder and takes more time. After a little over 4 hours, it is starting to look a lot better:
The run was stopped after 13 hours. Here is final screenshot:
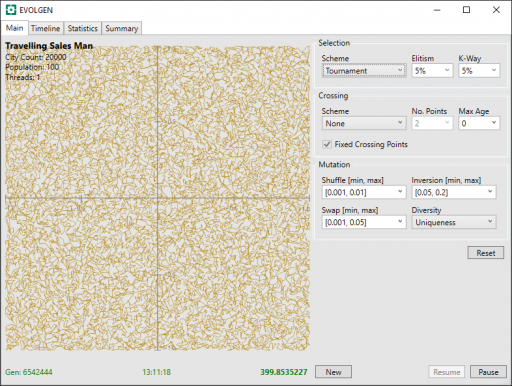
During the run, the tour distance has descreased from above 20558 to 400. Progress was still being made when the run was stopped.
Closing Remarks
Threads
A single thread was used in the test runs above. While EVOLGEN is designed to utilise multiple threads, this only comes to be an advantage when genome evaluation (i.e. calculating the route distance) is expensive. It was found, however, that route distance calculation was very fast. Employing multiple threads invoked thread synchronization which a single thread did not suffer from. The result was that there was little difference in performance when employing 8 threads instead of 1.
Inversion and Other Parameters
Working parameter values were found by trial and error. It was found, for example, that there was little difference in performance between a population 100 individuals and 200.
Note that, with EVOLGEN, mutation parameter values are chosen according to a population diversity measure within a specified [min, max] range. It was suprising that a relatively high inversion probability had a beneficial effect, with the value chosen in the range [0.01, 0.2]. It was suspected, given that both elitism and no crossover were employed, that the shuffle and swap mutation operations would benefit from similarly high values. However, it was found that increasing them actually had a detrimental effect. Setting the to zero, however, does not work either as the developing population then lacks a source of “entropy”.
The interplay between parameter values is complex. Without crossover and with low mutation values, many population members will go unmodified into the next generation. As stated, EVOLGEN does not perform unnecessary re-evalution. Generation processing in this scenerio is performed very quickly. Consequently, adjusting a parameter may yield improvements on a per generation basis, but generations may take longer to process. Therefore results, on a time basis, may seem counter intuitive.
As always, a more in-depth analysis may yield further answers.
Late update: It was found that setting K-Way to 15%, Elitism to 1, and Max Age to 100 yields possible performance improvement.
Improving Performance
EVOLGEN implements a number of technique for performance. For example, it does not re-evaluate genome sequence once evaluation has been performed and equality testing is very fast. However, a number of further ideas come to mind for improving performance with large city tours.
The area could be broken down into a grid of, say 6 x 6 cells with approximately 555 locations in each. Each of these cells could be subject of independent GA populations, with searching conducted concurrently. The optimized cells could then be stitched together and final search performed on the overall area. It is envisaged that this may yield a good/satisfactory result in a time frame of one to several minutes. However, futher progress would be computationally expensive.
Another possibility is to utilize parallel populations of the entire area, with “cross fertilization” occurring periodically. This approach has been subject of other studies, as described here [6].
Finally, the shuffle and swap operations are partial mutations operations. In these, two points are chosen at random and mutation occurs at or between these points respectively. However, with genomes that have undergone significant optimization, such operations are likely to be destructive. It is possible that new “localised” operations, in which the points are chosen closer together, may yield benefits.
References
[1] https://www.universetoday.com/36302/atoms-in-the-universe/
[2] GeneticSharp: https://github.com/giacomelli/GeneticSharp/tree/2.6.0
[3] Simulated annealing algorithm: https://github.com/forsythe/travelling-salesman
[4] Partially matched crossover: https://rubicite.com/Tutorials/GeneticAlgorithms/CrossoverOperators/PMXCrossoverOperator.aspx
[5] Order 1 crossover: https://rubicite.com/Tutorials/GeneticAlgorithms/CrossoverOperators/Order1CrossoverOperator.aspx
[6] Parallel and Distributed Genetic Algorithms: https://towardsdatascience.com/parallel-and-distributed-genetic-algorithms-1ed2e76866e3

